BioInformatics Tool for Infrastructure Automation (BiTIA) CLI utility
This is the cli utility of the BiTIA project. The other component is
bitia-runner. Most users only need the cli client bitia to submit jobs.
Install it using pip.
python -m pip install bitia --user
python -m bitia --helpConfiguration
TODO: Order of searching configuration file.
Unix like systems
./bitia.toml~/.bitia.toml$HOME/.config/bitia.toml/etc/bitia.toml
Windows
bitia.toml%APPDATA%\bitia.toml%PROGRAMDATA%\bitia.toml
BiTIA runner
If you are self-hosting the BiTIA server, you need bitia-runner as well. See
the documents of bitia-runner for more details. TODO.
EXAMPLES
- Using samtools to view a fastq file with same name as provided in url
| NORMAL PIPELINE EXECUTION CODE | BITIA EXECUTION CODE |
|---|---|
 |
 |
- Using samtools to view a fastq file with different file name as provided in url
| NORMAL PIPELINE EXECUTION CODE | BITIA EXECUTION CODE |
|---|---|
 |
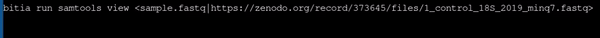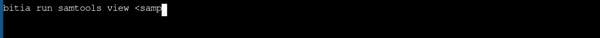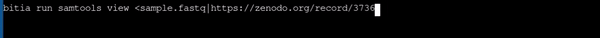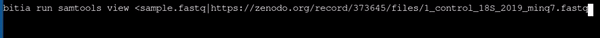 |
- Using Fastqc to do Quality check on a fastq file with same file name as provided
| NORMAL PIPELINE EXECUTION CODE | BITIA EXECUTION CODE |
|---|---|
 |
 |